Working with epidemiological data¶
The outbreak_data package contains endpoints that return epidemiological data on SARS-CoV-2. We can then visualize how SARS-CoV-2 is affecting countries around the world (with the help of one of the many plotting packages available for Python).
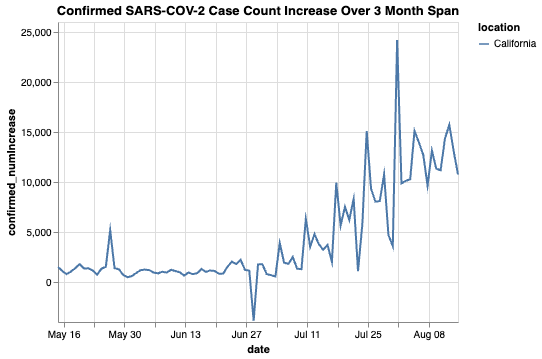
For example, we can look at the pattern of infection counts in California during a specific point in time:
# Perform authentication
from outbreak_data import authenticate_user
authenticate_user.authenticate_new_user()
# Import outbreak_data package
from outbreak_data import outbreak_data as od
import pandas as pd
# Get the number of increased cases from the previous day in California
counts_ca = od.cases_by_location('USA_US-CA')
# Formatting for graph
counts_ca= counts_ca.rename(columns={"admin1": "location"})
# Sort info by date and search within a date range
counts_ca = counts_ca.sort_values(by = "date")
counts_ca = counts_ca.loc[counts_ca["date"].between("2021-05-15", "2021-08-15")]
print(counts_ca)
#Import visual package of choice
import altair as alt
#Graph!
alt.Chart(xbb_mutations, title = "Daily ORF1a:K47R AND S:T19I Prevalence of Lineage XBB").mark_line().encode(
x='date:T',
y=alt.Y('proportion (%):Q'),
color = 'mutations:N')
Output¶
_id _score location \
621 USA_California_None2021-05-15 8.418888 California
622 USA_California_None2021-05-16 8.418888 California
623 USA_California_None2021-05-17 8.418888 California
624 USA_California_None2021-05-18 8.418888 California
625 USA_California_None2021-05-19 8.418888 California
.. ... ... ...
166 USA_California_None2021-08-11 8.419768 California
644 USA_California_None2021-08-12 8.418888 California
413 USA_California_None2021-08-13 8.418888 California
167 USA_California_None2021-08-14 8.419768 California
414 USA_California_None2021-08-15 8.418888 California
confirmed_numIncrease date
621 1504 2021-05-15
622 1087 2021-05-16
623 793 2021-05-17
624 1054 2021-05-18
625 1400 2021-05-19
.. ... ...
166 11164 2021-08-11
644 14356 2021-08-12
413 15707 2021-08-13
167 13100 2021-08-14
414 10744 2021-08-15
[93 rows x 5 columns]
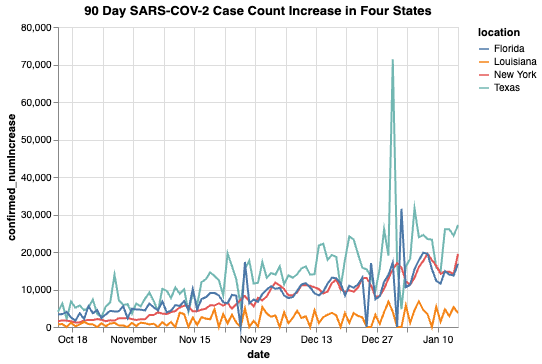
We can also do the same analysis over multiple locations and visualize them all at once:
counts_ca = od.cases_by_location('USA_US-NY')
counts_ny = od.cases_by_location('USA_US-TX')
counts_fl = od.cases_by_location('USA_US-LA')
counts_wa = od.cases_by_location('USA_US-FL')
state_count = pd.concat([counts_ca, counts_ny, counts_fl, counts_wa])
state_count = state_count.rename(columns={"admin1": "location"})
state_count = state_count.sort_values(by = "date")
state_count = state_count.loc[state_count["date"].between("2020-10-15", "2021-01-15")]
#Graph it!
alt.Chart(state_count, title = " 90 Day SARS-COV-2 Case Count Increase in Four States").mark_line().encode(
x='date:T',
y=alt.Y('confirmed_numIncrease:Q'),
color = 'location:N')
Note
The Vega-Altair visualization package is used for demonstration purposes. However, any Python visual package can be used to create graphical representations of the data.